CoRAL
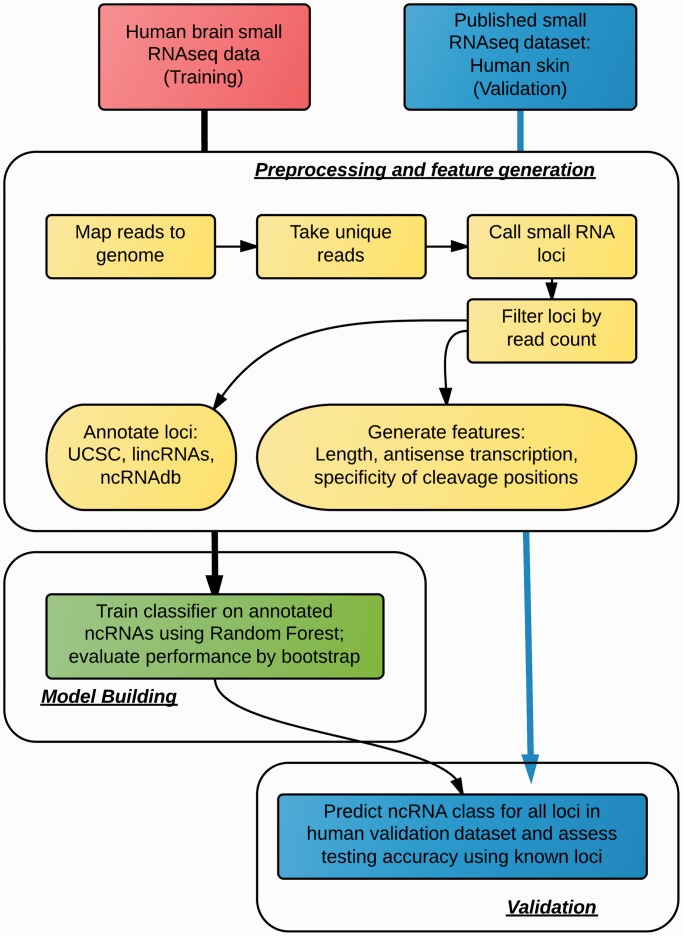
The surprising observation that virtually the entire human genome is transcribed means we know little about the function of many emerging classes of RNAs, except their astounding diversities. Traditional RNA function prediction methods rely on sequence or alignment information, which are limited in their abilities to classify the various collections of non-coding RNAs (ncRNAs). To address this, we developed Classification of RNAs by Analysis of Length (CoRAL), a machine learning-based approach for classification of RNA molecules. CoRAL uses biologically interpretable features including fragment length and cleavage specificity to distinguish between different ncRNA populations. We evaluated CoRAL using genome-wide small RNA sequencing data sets from four human tissue types and were able to classify six different types of RNAs with ∼80% cross-validation accuracy. Analysis by CoRAL revealed that microRNAs, small nucleolar and transposon-derived RNAs are highly discernible and consistent across all human tissue types assessed, whereas long intergenic ncRNAs, small cytoplasmic RNAs and small nuclear RNAs show less consistent patterns. The ability to reliably annotate loci across tissue types demonstrates the potential of CoRAL to characterize ncRNAs using small RNA sequencing data in less well-characterized organisms.