Understanding the genetic architecture of Alzheimer’s Disease and Related Dementias
Explore the ADSP Dataset through the NIAGADS Data Discovery Portal
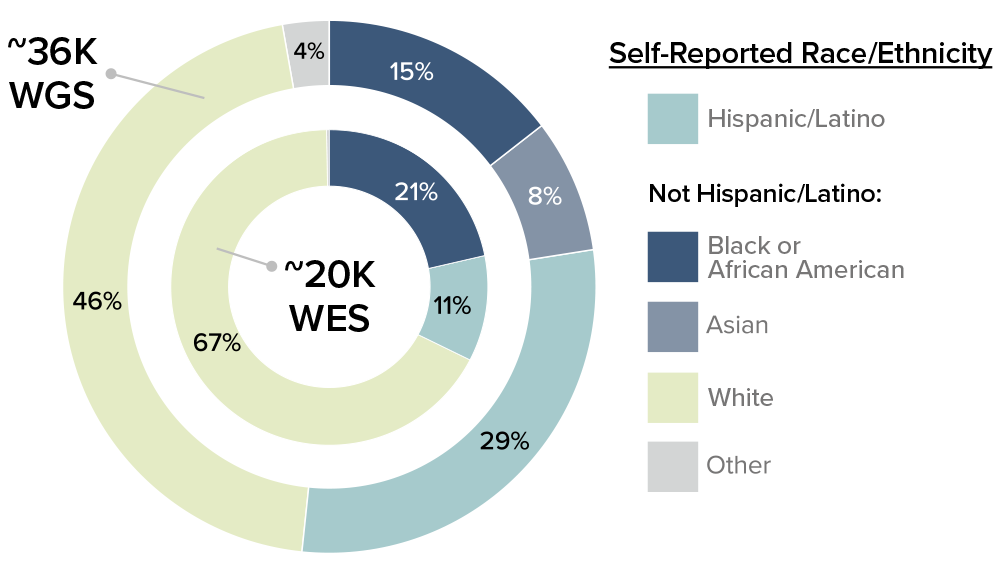
Powered by Gen3, the Data Discovery Portal, now in beta allows researchers to view summary information about cohorts, phenotypes, and available genetic data for the ADSP.
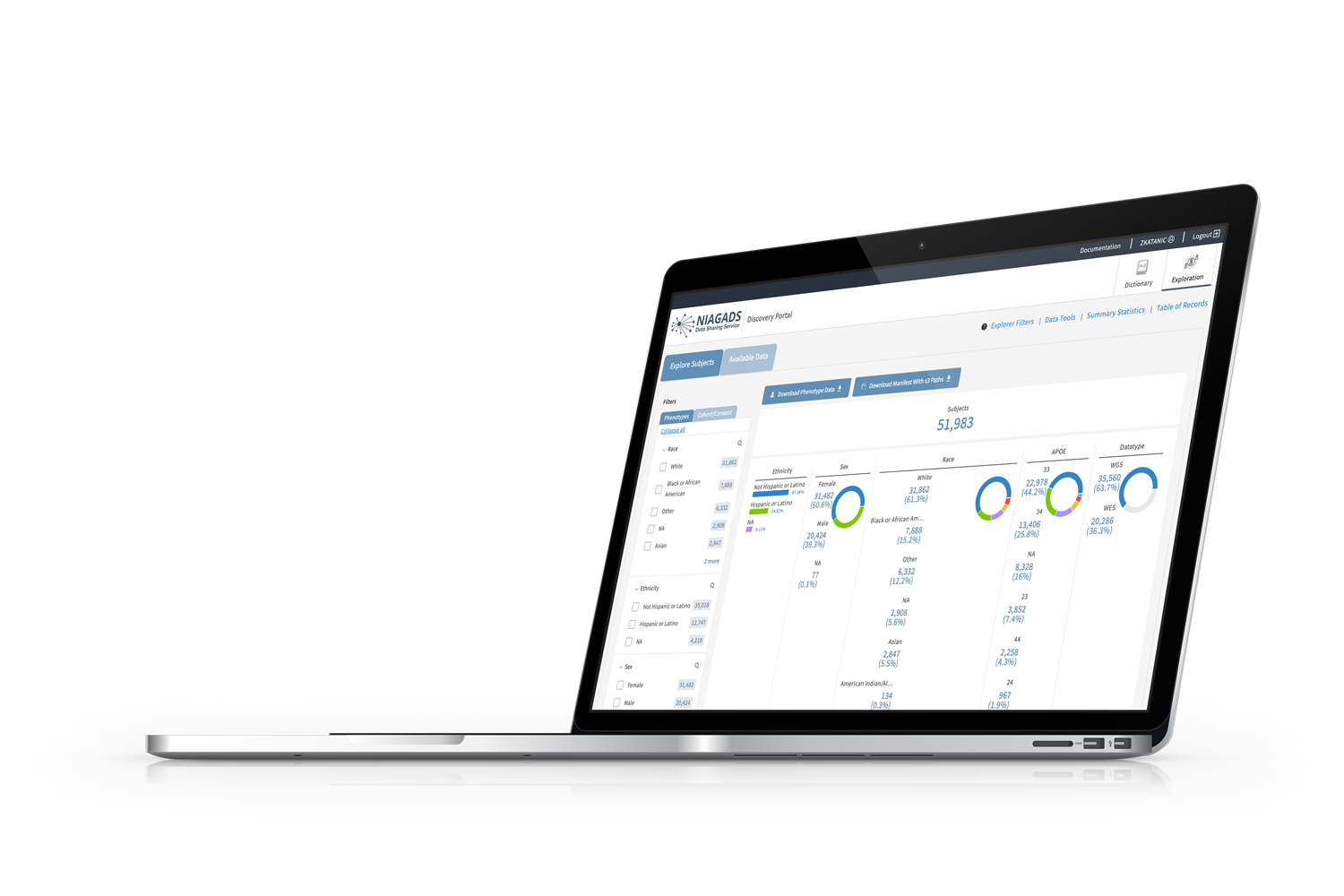
Available Data
For detailed information about the ADSP dataset including release notes and file manifests, or to apply for access to the dataset, go to the ADSP Umbrella Dataset page on NIAGADS DSS by clicking the button below.
*ADSP data is shared via NIAGADS DSS
Latest News
-
March 18, 2024
Creating the Largest Joint-Called Whole Exome Sequencing (WES) Dataset for Alzheimer’s Disease (AD)
Over the last 20 years, the technology for DNA sequencing has rapidly evolved, making it possible to sequence individual whole genomes on a large scale. But when trying to find the underlying genetic causes of disease, every... -
February 9, 2024
15 Genotyping Array Datasets From the Alzheimer’s Disease Research Centers (ADRC) Now Available!
We are thrilled to announce the release of 15 much-anticipated genotyping array datasets from participants recruited from the Alzheimer’s Disease Research Centers (ADRC)! These datasets, ADC1 through ADC15, are now available on the NIAGADS Data Sharing Service... -
January 5, 2024
Introducing hipFG: A high-throughput harmonization and integration pipeline for genomic annotations, QTLs, and chromatin interactions
A major challenge facing researchers today is the cleanup and harmonization of data for use in their work. As datasets get larger and the types of outputs continue to grow, this problem will continue to grow exponentially. Manually...